Currently, the pSSdb database contains pre-computed annotations/predictions for proteins from the following databases:
- UniProt (UniProtKB/Swiss-Prot, UniProtKB/TrEMBL; UniProt/UniParc version 2023_05; 578M protein sequences)
- AlphaFold database (215M protein sequences, >90k proteomes)
- ESMatlas (772M protein sequences)
- NCBI BLAST nr (non-redundant) (596M protein sequences)
- BFD (2.2B protein sequences)
- Uniclust (367M protein sequences)
- PDB70 (112M protein sequences, as used in AlphaFold2)
- MGnify (Full version, 3.0B protein sequences, and clustered version, 624M protein sequences, as used in AlphaFold2 are suported)
- KMAP (310M protein sequences)
- FESNov (400M protein sequences)
- GMGC (966M protein sequences)
- JGI_IMG (459M protein sequences)
Counter: 7,005,354,422 non-redundant protein sequences
The coverage statistics of pSSdb secondary structure predictions
(number of protein sequences in millions)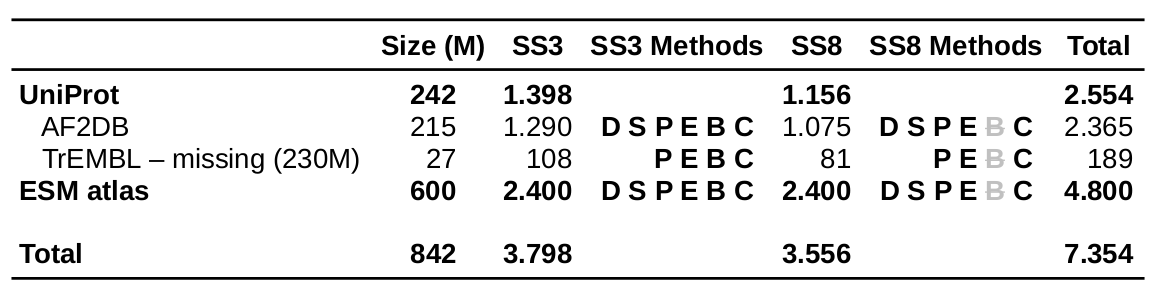
Methods DSSP and STRIDE are based on 3D structure models via AlphaFold2 (UniProt section) and ESMfold (ESM atlas section) respectively. For ProtBert (B) model only SS3 level of predictions are available.