What is protein secondary structure?
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure.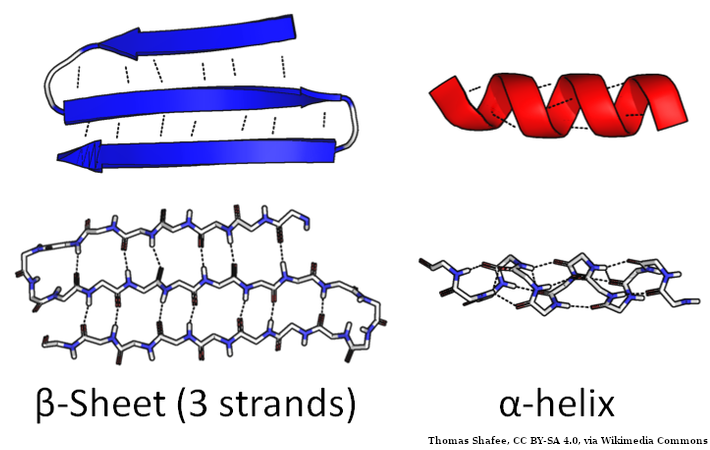
Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. For more details see Wikipedia article.
What is the primary UID in the pSSdb database?
The primary UID used in the pSSdb database is the protein sequence itself. This means that if the same sequence is present in multiple organisms, some annotations or links to external databases (such as UniProt or Taxonomy) may not be completely accurate.Therefore, while you can perform searches using UniProt, ESM Atlas, or AlphaFold UIDs, the most accurate and reliable results will be obtained by using the protein sequence as your query.
Example input sequence:
MVIHFSNKPAKYTPNTTVAFLALVDGAEVECEISVEALEDHFDAPSMQGVDLVAAFEAHRTQIEAVARVKLPQRLPAGRCLLISDYF
What are internal UIDs and how are they generated?
Each database implements its own identifiers for sequences, including the pSSdb database. Additionally, when combining data from multiple sources, there is a need to merge and remove redundant sequences. In pSSdb, we introduce identifiers that are calculated directly from the protein sequence using 64-bit hashes generated by the blake2b algorithm. The calculation process is as follows:1) Initialize the blake2b hash function with a digest size of 32, a person parameter of b'protein', and a salt parameter of b'protein'.
2) Update the hash function with the UTF-8 encoded protein sequence.
3) Obtain the blake2b UID by retrieving the hexadecimal digest of the hash function.
Python code:
h = blake2b(digest_size=32, person=b'protein', salt=b'protein' )
h.update(seq.encode('utf-8'))
blake2b_uid = h.hexdigest()
For example, given the protein sequence:MSNVGIVIVSHSPLVAEGTADMVRQMVGEEVPLAWCGGNGHGGLGTNVEAIMGAIDKAWSEAGVAILVDLGGAETNSEMAVEMIGEPRAHKIIVCNAPIVEGAVMAATEASGGASLREVVATAHELSPS
The corresponding blake2b UID is:
c26000d94289fb5eefbaac83b6928c21bb2edf51dcbd9e06cf6925d34b006c35
One important consideration with this approach is the risk of collisions, where two different sequences produce the same UID. However, even with billions of protein sequences and 64-bit hashing, the risk of collision is sufficiently low, less than 1 in 2100. For more detailed information, please refer to the provided link.
What is ss3-level secondary structure prediction?
The secondary structure of proteins can be summarized using three main elements represented by corresponding letters: H (helix), E (strand), - (coil). For more details see DSSP wikipedia article here.Below, you can find an example of ss3-level prediction:
3-letter alphabet: H (helix), E (strand), - (coil)
1.........11........21........31........41........51........61........71........81.....
MVIHFSNKPAKYTPNTTVAFLALVDGAEVECEISVEALEDHFDAPSMQGVDLVAAFEAHRTQIEAVARVKLPQRLPAGRCLLISDYF
BFD -EEEE-----------EEEEEEEE--EEEEEEEEHHHHH------------HHHHHHHHHHHHHHHHEEE---------EEEE----
bioemb -EEEE-----------EEEEEEEE---EEEEEEEEE--H------------HHHHHHHHHHHHHHHEEEE--------EEEEEE---
DSSP --EEE-----EEE---EEEEEEEE--EEEEEEEEHHHHHHHH------HHHHHHHHHH-HHHHHHHHHHHHHHHHHH--EEE-HHH-
STRIDE --EEE-----EEE---EEEEEEEE--EEEEEEEEHHHHHHHH------HHHHHHHHHHHHHHHHHHHHHHHHHHHHH--EEE-HHH-
SSE-PSSM EEEEEE-------------HH-------EEHHHHHHHHH------------HHHHH--HHHHH-HHH-EE---------EEEEEH--
Consensus -EEEE-----------EEEEEEEE--EEEEEEEEHHHHH------------HHHHHHHHHHHHHHHHHEE---------EEEEHH--
SS3 conf 648775778844488767777776884677777766657344777778443787887767787677533344444446577733437
MVIHFSNKPAKYTPNTTVAFLALVDGAEVECEISVEALEDHFDAPSMQGVDLVAAFEAHRTQIEAVARVKLPQRLPAGRCLLISDYF
BFD -EEEE-----------EEEEEEEE--EEEEEEEEHHHHH------------HHHHHHHHHHHHHHHHEEE---------EEEE----
bioemb -EEEE-----------EEEEEEEE---EEEEEEEEE--H------------HHHHHHHHHHHHHHHEEEE--------EEEEEE---
DSSP --EEE-----EEE---EEEEEEEE--EEEEEEEEHHHHHHHH------HHHHHHHHHH-HHHHHHHHHHHHHHHHHH--EEE-HHH-
STRIDE --EEE-----EEE---EEEEEEEE--EEEEEEEEHHHHHHHH------HHHHHHHHHHHHHHHHHHHHHHHHHHHHH--EEE-HHH-
SSE-PSSM EEEEEE-------------HH-------EEHHHHHHHHH------------HHHHH--HHHHH-HHH-EE---------EEEEEH--
Consensus -EEEE-----------EEEEEEEE--EEEEEEEEHHHHH------------HHHHHHHHHHHHHHHHHEE---------EEEEHH--
SS3 conf 648775778844488767777776884677777766657344777778443787887767787677533344444446577733437
Note that for simplicity confidence scores are shown only for consensus prediction. For individual scores of other methods download the raw output in fasta format.
What is ss8-level secondary structure prediction?
Protein secondary structure can be definied at more precise level where we use 8 elements: G (310 helix), H (α helix), I (Π helix), B (β bridge), E (β bulge), T (turn), S (curvature), - (other). For more details see DSSP wikipedia article here.Below, you can find an example of ss8-level prediction:
1.........11........21........31........41........51........61........71........81.....
MVIHFSNKPAKYTPNTTVAFLALVDGAEVECEISVEALEDHFDAPSMQGVDLVAAFEAHRTQIEAVARVKLPQRLPAGRCLLISDYF
bioemb --EEE--------TT-EEEEEEEETTEEEEEEEEEEE-GTT----T-TT--HHHHHHHHHHHHHHHEEEE--S---TTEEEEEEE--
DSSP --EEE-----EEETTTEEEEEEEETTEEEEEEEEHHHHHHHH--S-SSHHHHHHHHHHTHHHHHHHHHHHHHHHGGGT-EEE-GGG-
STRIDE --EEE-----EEETTTEEEEEEEETTEEEEEEEEHHHHHHHH------HHHHHHHHHHHHHHHHHHHHHHHHHHGGG--EEE-GGG-
SSE-PSSM EEEEEEHS-T-ES----HHHHH-HTT--EEHHHHHHHHHHHT----SS--HHHHHHHHHHHHHHHHHEEE---E--TS-EEEEEHSS
Consensus --EEE-----EEETTTEEEEEEEETTEEEEEEEEHHHHHHHH-----SHHHHHHHHHHHHHHHHHHHHHHHHHHGGGT-EEE-GGG-
SS8 conf_ 567776567645466466666666776677666655555554664532445777777757777777544444433333477744446
MVIHFSNKPAKYTPNTTVAFLALVDGAEVECEISVEALEDHFDAPSMQGVDLVAAFEAHRTQIEAVARVKLPQRLPAGRCLLISDYF
bioemb --EEE--------TT-EEEEEEEETTEEEEEEEEEEE-GTT----T-TT--HHHHHHHHHHHHHHHEEEE--S---TTEEEEEEE--
DSSP --EEE-----EEETTTEEEEEEEETTEEEEEEEEHHHHHHHH--S-SSHHHHHHHHHHTHHHHHHHHHHHHHHHGGGT-EEE-GGG-
STRIDE --EEE-----EEETTTEEEEEEEETTEEEEEEEEHHHHHHHH------HHHHHHHHHHHHHHHHHHHHHHHHHHGGG--EEE-GGG-
SSE-PSSM EEEEEEHS-T-ES----HHHHH-HTT--EEHHHHHHHHHHHT----SS--HHHHHHHHHHHHHHHHHEEE---E--TS-EEEEEHSS
Consensus --EEE-----EEETTTEEEEEEEETTEEEEEEEEHHHHHHHH-----SHHHHHHHHHHHHHHHHHHHHHHHHHHGGGT-EEE-GGG-
SS8 conf_ 567776567645466466666666776677666655555554664532445777777757777777544444433333477744446
How to interpret the confidence of secondary structure predictions?
At web page of the database we use 10 levels and following coloring:Model Confidence (SS3 conf and SS8 conf lines):
Very high (scores 7-9)
Confident (scores 5-6)
Low (scores 3-4)
Very low (scores 0-2)
Additinally, when you dowload raw output in pseudo-fasta format you can obtain more exact confidence levels including confidence scores for individual methods.
In most cases, you are able to estimate the prediction accuracy. This can be some kind of score (usually normalized to 0-1 or 0-100 range) or some third-party metrics (e.g. quality of the model).
In pSSdb, we use 50 bins for confidence scores (equivalent of 2% threshold). This is done mostly from technical reasons i.e. the way how the scores are encoded.
ENCODING CONFIDENCE SCORES (for raw download files)
To make the coding simple we are using ASCII letters to encode 50 thresholds:
#import string; string.ascii_letters[:50] alphabet = 'abcdefghijklmnopqrstuvwxyzABCDEFGHIJKLMNOPQRSTUVWX' score = 0.7889 #score must be normalized in the range [0, 1] alphabet_index = int(round(math.floor(score*50),0)) if alphabet_index==50: alphabet_index=49 encoded_score = alphabet[alphabet_index] print(alphabet_index, encoded_score) 39 NDECODING
decoded_score = alphabet.index(encoded_score) / 50.0 print(decoded_score) 0.78
Thus going back to our example:
1.........11........21........31........41........51........61........71........81..... 51e01e8bce3aacee923f81ef07006ec7352a2436c4441736c0220176e3dc051d|AF-A0A4R5KZE2-F1|tax-id 2546446 MVIHFSNKPAKYTPNTTVAFLALVDGAEVECEISVEALEDHFDAPSMQGVDLVAAFEAHRTQIEAVARVKLPQRLPAGRCLLISDYF sequence -EEEE-----------EEEEEEEE--EEEEEEEEHHHHH------------HHHHHHHHHHHHHHHHEEE---------EEEE---- ss3 WGPPFDMRRPLKQTTKHTVWWWVTWWJUUVVUROLPQEAzEMQTONOPQJGVWWWWWVPRQRLIGBvyBEKQQMSTTOFFLLttyFX scoresAs you can see, the scores are usually uppercase letters, thus you can consider the prediction reliable, less reliable fragment (but steal resonable) is AR (positions 67-68) within ...HE... ss3 fragment (swithc from helix to beta) and scores are 'vy'.
How the consensus is calculated?
The consensus is calculated based on predictions from all available methods, taking into account the prediction scores from each method. For the DSSP and STRIDE methods, confidence scores are derived from the model's accuracy at the individual residue level (using pLDDT if based on the AlphaFold model, or pTM & pLDDT for ESM Atlas models). If a method provides prediction confidence scores (such as SSE-PSSM), those scores are instead used in the consensus calculation.
What protein databases are covered by pSSdb?
Currently, the database contains annotation for proteins from the following databases:
- UniProt (UniProtKB/Swiss-Prot, UniProtKB/TrEMBL; UniProt/UniParc version 2023_04; 543M protein sequences)
- AlphaFold database (215M protein sequences, >90k proteomes)
- ESMatlas (772M protein sequences)
- NCBI BLAST nr (non-redundant) (596M protein sequences)
- BFD (2.2B protein sequences)
- Uniclust (367M protein sequences)
- PDB70 (112M protein sequences, as used in AlphaFold2)
- MGnify (Full version, 3.0B protein sequences, and clusterd version, 624M protein sequences, as used in AlphaFold2 are suported)
- KMAP (310M protein sequences)
- FESNov (400M protein sequences)
- GMGC (966M protein sequences)
- JGI_IMG (459M protein sequences)
What algorithms/programs are used for secondary structure prediction?
The prediction of secondary structure is done using the following programs/algorithms:- DSSP (ss3 and ss8 alphabet secondary structure annotations from 3D protein models from ESMatlas and AlphaFold databases)
- STRIDE (ss3 and ss8 alphabet secondary structure annotations from 3D protein models from ESMatlas and AlphaFold databases)
- SSE-PSSM (ss3 and ss3 sequence-based predictions)
- Bio Embeddings (seqvec model, ss3 and ss8 sequence-based predictions)
- ProtTrans (prot_bert_bfd_ss3 model, ss3 only sequence-based predictions)
pSSdb database is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP pSSdb: Protein Secondary Structure Database (submitted) | Contact: Lukasz P. Kozlowski |